cDNA Libraries
cDNA
Vectors
Biology
Vectors
In order to obtain access to this information, we must convert the unstable mRNA population into a DNA copy, or cDNA.
This enzyme reverse transcriptase allows us to use an RNA template to produce a double-stranded cDNA copy. Reverse transcriptase was discovered by H. Temin and D. Baltimore while studying retroviruses. Retroviruses contain an RNA genome which is converted to a DNA copy and integrated into the host genome during its replicative cycle. This is an interesting set of viruses including many tumor viruses and the AIDS virus HIV.
Reverse transcriptase is an RNA-dependant DNA polymerase. It utilizes RNA as a template, requires dNTPs and a primer (free 3' OH) to initiate DNA polymerization.
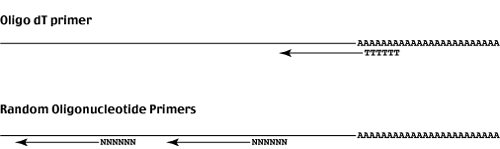
Two types of primer are commonly used.
Oligo-dT initiates priming from the 3' end of the mRNAs.
cDNAs primed with oligo-dT are enriched for mRNA 3' ends.
Random oligonucleotides can hybridize anywhere along the mRNA sequence and prime cDNA synthesis. Randomly primed cDNAs are distributed along the length of the template and are therefore more representitive of the mRNA population.
mRNAs are typically short (as compared to the genome) - most mRNAs are under 6 kb in length and only rare mRNA exceed 10 kb in length.
This small size means that both plasmid and phage insertion vectors are appropriate for the construction of cDNA libraries
(in contrast to genomic libraries where phage substitution vectors are preferred).
In our discussion of genomic libraries, we focused on complete coverage of genome.
Random genomic fragments were generated by partial digestion with a frequent cutting enzyme.
The resulting random shotgun library contains multiple overlapping clones that cover the complete genome sequence.
cDNA libraries are a little different.
Here each bacterial transformant or packaged phage represents a unique mRNA molecule.
Recombinants containing the same DNA sequence represent different template molecules present in the e original mRNA population.
For example, there are 100,000 mRNA molecules in the cell at a given time
10% of them are some highly expressed mRNA (lets say actin mRNAs)
then in a primary cDNA library consisting of 100,000 clones,
10% of them (10,000) will be actin cDNAs.
Or you could just screen a couple of hundered cDNAs and still find actin cDNAs.
Well, thats very nice if you want to study actin.
What if you want to study some rare transcription factor that is only expressed at low levels.
In your 100,000 mRNAs, there may be only 10 mRNAs encoding your transcription factor.
Now you would have to screen your entire library of 100,000 clones.
If your transcript was only expressed for a short time at low levels, it might be present at even lower levels.
The majority of recombinant phage in a standard cDNA library carry highly expressed sequences.
Rare mRNAs are hard to find unless your library is very large (number of recombinants - should contain more than 106 independant recombinants to cover a mRNA population of 100,000 transcripts with 99% probability).
The alternative to screening increasing numbers of independant recombinants is to 'normalize' the library utilizing hybidization kinetics as we discussed previously.
Normalized libraries contain fewer copies of highly expressed mRNAs (removed on hybridization) and more copies of rare transcripts (in a relative sense).
go to
Hybridization Technology