Other Vector Systems
Vector
Systems
Vectors
Biology
Vectors
A major thrust of vector development has been to create vectors that will handle larger foreign DNA inserts, aiming to reduce the number of recombinants it is necessary to look at in order to identify a specific DNA sequence.
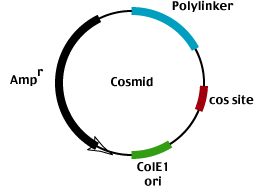
Cosmid vectors were among the first large insert cloning vehicles developed.
The vector replicates as a plasmid
(it contains a ColE1 origin of replication),
uses Ampr for positive selection and
employs lambda phage packaging to select for recombinant plasmids carrying foreign DNA inserts 45 KB in size.
Ligation of cosmid vector and foreign DNA fragments (SauIIIA partial digest fragments 45 kb in size) is similar to ligation into a lambda substitution vector.
The desired ligation product is a concatemer of
45 kb foreign DNA fragment and 5 kb cosmid vector sequences.
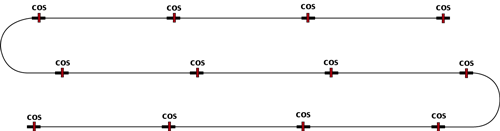
This concatemer is then packaged into viral particles (remember packaging is cos site to cos site) and these are used to infect E. coli where the cosmid vector replicates using the ColE1 orignin of replication. Phage packaging serves only to select for recombinant molecules and to transfer these long DNA molecues (50 kb total) into the bacterial host (50 kb fragments transform very inefficiently while phage infection is very efficient).
Recently, the need for prokaryotic vectors capable of replicating very large foreign DNA fragments
(> 1 MB in size) has produced a couple of additional prokaryotic vector systems.
One is based on another bacteriophage called P1.
P1 phage can carry in excess of 100 KB of foreign DNA but do not seem to have found wide acceptance.
A few years ago, a Bacterial Artificial Chromosome (BAC) vector was developed allowing foreign DNA fragements over 1 MB to be propagated in E. coli.
These BAC vectors replicate as low copy number plasmids (to minimize the demands on the host replication system) but how do we get such large DNA fragments into E. coli?
Standard methods for transforming E coli rely on the chemical preparation of the E coli to put them in a 'transformation competent' state.
This typically involves incubating the bacteria in the presence of divalent cations (particularly Ca+2) at low temperature (4 oC) which puts the cell membrane into a 'semi-crystaline state. Addition of DNA to these 'competent cells' allow the DNA to bind to the semi-crystaline membrane. Heat shocking the cells (42 oC) remobilizes the membrane and allows the DNA to cross the membrane. This transit across the membrane is sensitive to DNA length - small plasmids cross the membrane rapidly while larger molecules are unable to cross.
Getting very large molecules across the cell membrane relies on an alternative mechanism called electroporation. This involves mixing DNA with E coli cells and then exposing the mixture to a high-voltage pulse. The high voltage induces massive membrane rearrangements and the coincident uptake of DNA which is independant of size. BACs have been an important tool for the human genome sequencing project.
Numerous vector systems have been developed for use in eukaryotic systems.
The most common of these are often called 'shuttle vectors' as they replicate in both prokaryotic and eukaryotic hosts. In general, DNA manipulations and characterizations are done in prokaryotic systems, and then the manipulated DNA is reintroduced to eukaryotic systems for functional analysis.
Yeast Vectors
Shuttle vectors were first developed for transfering DNA fragments between E coli and S cerevisiae. These vectors contain an antibiotic resistance gene for positive selection in E coli, and E coli origin of replication (ColE1 ori), and a polylinker in the lacZ alpha-complementing fragment for insertional inactivation by the foreign DNA insert. In addition, the vector contains a yeast specific origin of replication (derived from the naturally occuring yeast plasmid called the 2 um circle - 2um ori)
and amino-acid or N-base biosynthsis enzymes for positive selection in yeast (HIS, LEU and ADE genes)
Mammalian Vectors
Shuttle vectors have also been developed for use in mammalian tissue culture.
Like the yeast shuttle vectors, mammalian shuttle vectors require sequeces enabling them to replicate in mammalian tissue culture. These eukaryotic origins of replication are typically derived from well characterized mammalian viruses - most commonly SV-40 (SV-40 ori and Large T antigen system) or Epstein-Barr virus (mononucleosis). Both systems allow the vector to replicate within the host cell as a plasmid without integrating into the genome. In addition to the origin of replication, these shuttle vectors also carry antibiotic resistance genes which function in eukaryotic cells (ie neomycin (G418) resistance, hygromycin resistance, methotrexate resistance etc).
Finally, artificial chromosome vectors have been developed for use in yeast (YACs) and are being developed for use in mammalian cells (MACs).
These eukaryotic vectors contain telomeric sequences (eukaryotic chromsomes are linear not circular molecules) and centromeres to ensure appropriate segregation of the artificial chromosomes as well as selectable markers. Such vector systems may gain in importance as we move towards manipulating the eukaryotic genome.
go to
Libraries