Transcription II
The figure below attempts to give you some picture of what the polymerase - template - transcript 'looks like' during the elongation phase of transcription. The relative sizes of the template and polymerase (and some of the internal features of the polymerase itself) are pretty much as illustrated.
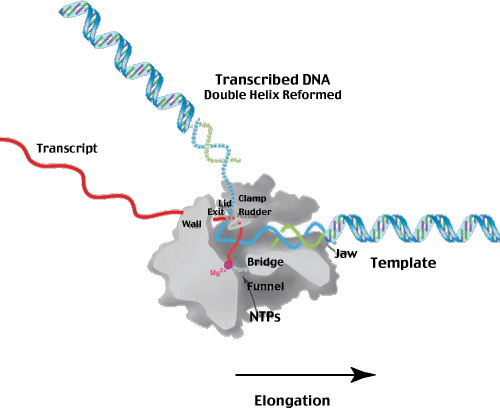
Hopfully, the illustration will help you visualize how the polymerase distorts the helix - facilitating unwinding at the active site. Note that the active site contains a Mg+2 ion - to stabilize the charges on the NTP substrates. You might also consider how the DNA-RNA hybrid formed by synthesis is broken to allow the original DNA-DNA double helix to reforn as it exits the polymerase.
The elongation phase is (at least in prokaryotes) essentially 'unregulated'. The synthesis of the message proceeds at the maximum rate permitted by enzyme kinetics and NTP substrate concentration.
In eukaryotes there is some indication that the rate of elongation may be a regulatory target. Modifications of the nucleosome structure can be envisioned which might make subsets particularly good or bad elongation templates.
Lets just say the jury is still out on this one...
In E. coli, transcription termination occurs by two general mechanisms.
The first mechanism is called rho dependant.
Rho is a protein - a termination factor (analogous to sigma - an initiation factor). Rho recognizes a specific DNA sequence and interacts with the transcription machinery to terminate transcription at that site.
The second mechanism is called rho - independant.
That is to say, it acts independantly of rho factor - or any other termination factor. The rho-independant signal is actually encoded in the transcript itself.
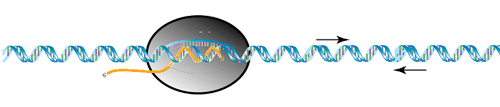
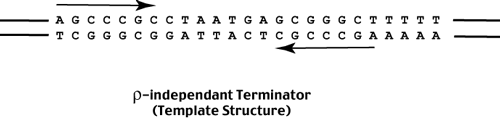
As the RNA polymerase continues synthesis it approaches a region of 'dyad symmetry' - this region is indicated by the two arrows above and below the template helix shown above. The sequence of the DNA is also shown. As the RNA polymerase transcribes across this sequence, the transcript forms a hair-pin loop as shown below. This hair-pin structure IS the rho-independant terminator. The hair-pin physically trips a switch near the exit channel which causes the RNA polymerase to open - release the template and transcript - and terminate transcription.

