Regulation of Transcription
The lac Operon Model
Sequence Specific DNA Binding Proteins
We previously divided the transcription process into three stages - initiation, elongation and termination.
Of these three stages, the initiation process is the target of most regulatory mechanisms.
Recall our previous discussion of the structure of the lac operon
The lac operon encodes the biochemical pathway enabling E coli to utilize lactose as a Carbon Source.
If there is no lactose present in the environment, E. coli shuts down transcription of this operon as a way of saving energy (gene expression, by its very nature, requires the expenditure of energy).
How does E coli turn off transcription of the lac operon?
The work of F. Jacob and J. Monod (and others) in the 1960's on this problem provided us with an elegant explanation of transcriptional regulation which remains conceptually relevant 40 years later.
The turning off of lac operon expression is due to the constitutive expression of
the lac repressor - a sequence-specific DNA binding protein encoded by the E coli lac I gene.
Before we get into how the lac repressor affects the transcription process, lets examine the interaction between proteins and DNA in a bit more detail.
In our discussion of nucleosome structure we noted that the interaction of the Histone proteins with DNA involved electrostatic interactions between the basic (+ charged) residues of the Histone and the acidic (- charge) sugar-phosphate backbone. In the nucleosome, this interaction is independant of the nucleotide sequence of the DNA since it only invovles electrostatic interactions with the backbone.
Most Protein-DNA interactions involve an electrostatic interaction between the protein and the DNA backbone. These electrostatic interactions allow proteins to bind 'non-specifically' to the DNA molecule and facilitate the search for a specific nucleotide sequence to bind tightly to. The continuous sugar-phosphate backbone acts like a set of railroad tracks on which the protein scans for its binding site.
We have discussed the structure of the double-helix - the major groove of the helix is formed from the edges of the paired bases and its surface therefore contains sequence-specific information. The surface of the major groove is what a sequence-specific DNA binding protein searches to find its specific binding site. When the surface of the major groove matches the surface of the protein, the two mesh and the protein remains bound at that target site.
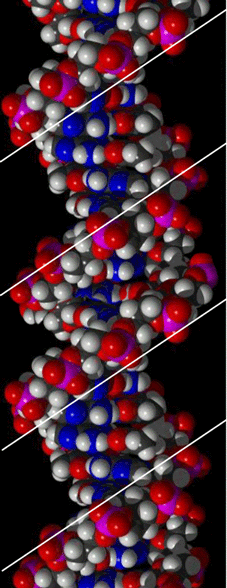
The Major Groove
These figures clearly show that the surface of the major groove is formed by the edges of the complementary Nitrogen Base pairs.
The width of the major groove is sufficient to accomodate a protein alpha-helix or Zn-finger. The surface of these sequence-specific DNA binding domains match the Major Groove surface at their specific target sites.
(see figure at the bottome of the page)
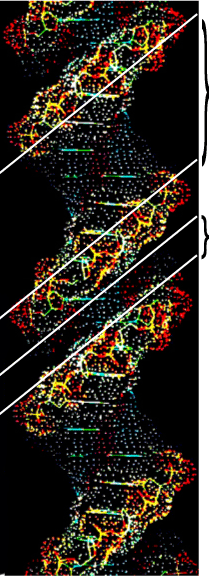
These figures also clearly show that the Minor Groove is too narrow to permit access to the protein DNA binding domains as discussed above for the Major Groove.
The figure below shows the binding of a prokaryotic repressor to its recognition site. Unfortunately, it isn't the lac repressor (I'm sure its out there somewhere but I just don't have the time to hunt it down). The protein is the lambda repressor (we'll talk about it a bit later) which regulates the transcription of the bacteriophage lambda genome.
I have tried to highlight the interaction between the sequence-specific DNA binding domain (an alpha-helix in this case) and the surface of the Major Groove using a green highlight.
The electrostatic interaction between the lambda repressor and the sugar-phosphate backbone are circled in white.
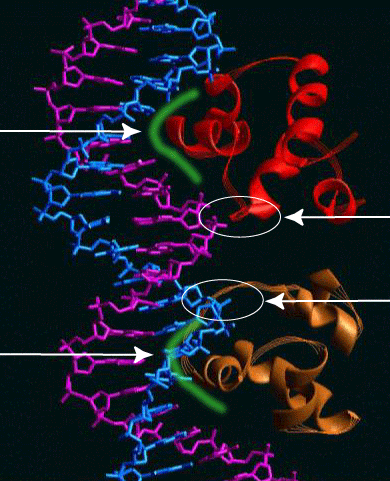
A couple of minor points -
The lambda repressor acts as a dimer - the two dimers are shown in red and 'bronze'.
The recognition sequence exhibits dyad symmetry - two opposites oriented repeats.
The two monomers bind to one another by
protein-protein interactions.
The contact with the sugar phosphate backbone spans the Minor Groove.
go to
Lac Operon Regulation