
Figure 8:
Characteristics of the B-form of the DNA Double Helix
When dehydrated, DNA takes on the right-handed A-form conformation in which the bases are tilted with respect to the long axis of the helix. The A-form is believed to be the conformation of DNA-RNA hybrids, and may represent the conformation of the template when being transcribed.
Additional conformations of the double helix have been described for specific sequences.
A left-handed Z-form has been described for sequences of very high GC content.
Regions of high AT content embedded in random sequence have been proposed to introduce a bend in the helix which can affect fragment migration through an agarose gel.

Figure 9: Comparison of A-, B- and Z-forms of the double helix
(arrows show tilt of bases relative to long axis of helix)
First, the H-bonds between the complementary bases hold the two strands together.
Note that the G:C content of the sequence will influence the overall stability of the helix
as G:C pairs share 3 H-bonds while A:T pairs only share two.
Hydrophobic interactions also play a large part in stabilizing the helix.
Since the aromatic rings N-bases are hydrophobic, these interactions keep the bases confined to the center of the helix where they are protected from interacting from the polar solvent.
Finally, the stacking of N-bases down the center of the helix adds additional stability.
The stacking energy is a combination of Van der Waals forces between the p-electron clouds of the aromatic rings (a hydrophobic interaction) and the interaction of the dipole groups.
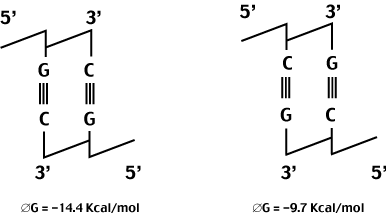
In general, the forces holding the helix together are not particularly sequence specific.
The stacking energy is an exception to this rule. Because of the way the bases are oriented down the center of the helix, the amount of p-overlap and dipole proximity will depend on the actual sequence.
An example of this sequence dependance is shown below.
go to
Chromatin Structure and DNA Packaging