Restriction Enzymes
Restriction enzymes have obscure sounding names which are directly derived from the bacterial strains from which they were first isolated. Most of the restriction enzymes we purchase commercially have been cloned and are now isolated from engineered E. coli expressing them - they still bear their original names however. Several examples of how these enzymes are named are shown below.
Providencia stuartii
1st enzyme isolated
Escherichia coli
strain RY103
1st enyzme isolated
Thermus aquaticus
1st enzyme isolated
Haemophilus influenzae
strain RD
3rd enzyme isolated
Restriction enzymes are classified on the basis of two fundamental properties.
First, whether the enzyme cuts at its recognition site, and second whether the restriction and modification activities are associated with the same or different complexes.
cuts at recognition site
restriction and modification activities part of the same complex.
cuts at recognition site
restriction and modification activities part of different complexes
cuts an arbitrary distance from recognition site
restriction and modification activities part of same complex.
Restriction endonucleases recognize specific sequences 4 to 8 base pairs long. These restriction sites are typically pallindromic - they read identically on both strands.
Mechanistically, restriction enzymes are believed to bind non-specifically to the sugar-phosphate backbone of the target molecule. The enzyme can then use the backbone as a track to scan the DNA for an appropriate sequence by probing for sequence-specific features in the major groove of the molecule. On finding such a site, the enzyme binds, and in the presence of Mg+2 undergoes a conformational change which kinks the helix and breaks the backbone of each strand to produce fragments with 5' phosphate and 3' hydroxyl groups.

If the enzyme cuts at the axis, blunt ends are produced.
If the enzyme cuts to the left of the axis, 5' overhangs are produced.
If the enyzme cuts to the right of the axis, 3' overhands are produced.
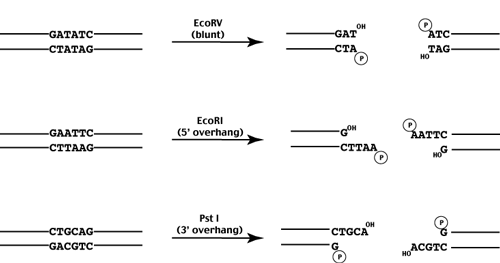
While each restriction enzyme recognizes and cleaves a specific sequence, a given recognition sequence may be recognized by multiple enzymes. Enyzmes which recognize the same sequence are called isoschizomers. Recently these have been further divided into two classes - isoschizomers and neoschizomers - which cleave the DNA at the same position or different positions respectively.

Frequency of Restriction Enzyme Sites
The frequency with which restriction sites occur in a random sequence can be simply calculated if the GC content of the random sequence is known. For each nucleotide position in the restriction site, determine the frequency with which that position is occupied by the appropriate base. Then multiply the frequencies together to obtain the frequency with which the complete site is observed.
4-mer sequence will occur
1/4 x 1/4 x 1/4 x 1/4
= 1/256 bp
6-mer sequence will occur
1/4 x 1/4 x 1/4 x 1/4 x 1/4 x 1/4
= 1/4096 bp
8-mer sequence will occur
1/4 x 1/4 x 1/4 x 1/4 x 1/4 x 1/4 x 1/4 x 1/4
= 1/65536 bp

2/10 x 3/10 x 3/10 x 2/10
= 1/278 bp
Similarly the 6-mer would cut
2/10 x 2/10 x 3/10 x 3/10 x 2/10 x 2/10
= 1/6944 bp
and the 8-mer would cut
2/10 x 2/10 x 2/10 x 2/10 x 2/10 x 2/10 x 2/10 x 2/10
= 1/390625 bp
Average fragment size and average frequency of restriction enzyme cutting depends on both the length and the GC bias of the recognition site as well as the GC bias of the genome being cut.
go to
Cloning Vectors