Hybidization Technology
Denaturing Nucleic Acids
1) H-Bonds between complementary bases
2) Stacking of the hydrophobic N-bases down the center of the helix
In opposition to these stabilizing interactions is the electrostatic repulsion of
the charged sugar-phosphate backbone.
There are two basic approaches to denaturing double-stranded DNA
- heating and chemical treatiment.
Chemical denaturants can be divided into three classes
Bases like NaOH raise the pH until the H+ shared between the N-base electronegative centers
( N-H and O= ) is stripped from the H-bond.
Loss of H-bonds between two complementary strands results in strand separation.
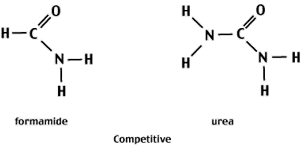
Compounds like urea and formaldehyde contain functional groups that can form H-bonds with the electronegative centers of the N-bases.
At high concentrations (8M urea or 70% formamide) of the denaturant, the competition for H-bonds favors interactions between the denaturant and the N-bases rather than between complementary bases. As a result, the two strands separate.
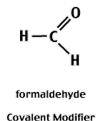
Reactive aldehydes like formaldehyde and glyoxal can covalently modify the electronegative centers of the N-bases and thereby block the formation of H-bonds between complementary bases.
Covalent modification is reversible.
Before leaving the topic of DNA denaturation, lets look a little more closely at
Heat Denaturation.
Consider what happens when we heat a nucleic acid solution - say the E. coli genome. To prepare the DNA for this experiment, we shear it up into small pieces (approximately 500 bp long) and heat it slowly while monitoring the A260.
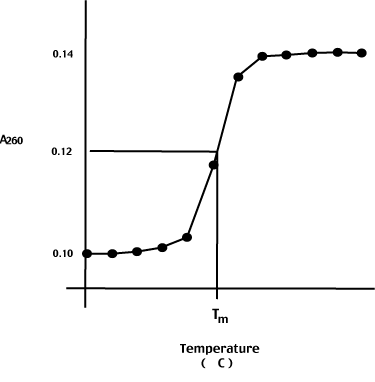
The initial A260 is stable until, over an interval of approximately 5 degrees C, the A260 suddenly increases by approximately 40%.
This increase in absorbance is referred to as the
Hyperchromic Shift.
The hyperchromic shift is due to the melting of the double helix into two single strands. The increased rotational freedom of the N-bases on strand separation accounts for the observed increase in absorbance.
The melting temperature, or Tm, is the temperature at the midpoint of the hyperchormic shift as shown to the left.
Three main factors affect the melting temperature.
The GC content of the nucleic acid sample.
This is due to the fact that AT base pairs share 2 H-bonds while GC base pairs share 3 H-bonds.
[salt]
Tm is sensitive to Na+ concentration.
Na+ acts to shield the negative charges of the sugar-phosophate backbone from interacting with one another. The repulsion between the negatively charged phosphate backbones is the major force destabilizing the double helix, therefore increasing Na+ concentration increases helix stability and decreasing Na+ concentration decreases helix stability.
DNA hybrid length
The longer the DNA hybrid is, the more H-bonds there are holding the two strands together. The longer the hybrid, the more H-bonds that must be simultaneously broken for the two strands to separate.
This is known as the 'zipper effect' after the (in)famous Canadian inventor Zippy. For our purposes we will only consider the two extremes of the zipper effect. For this course we will only consider the extemes of hybrid length - hybrids less than 50 bp (short) and those around 500 bp (long) in length.
For short DNA hybrids, the 'zipper effect' alters the formula to
Renaturation