Index
Plasmid Vectors I
Vectors
II
Enzymes
Vectors
I
Required for automous replication of the plasmid using the host's replication machinery.
Almost all commonly used plasmids are based on the ColE1 origin of replication (ori).
It is worth noting that bacterial origins of replication are tightly regulated.
While R factors are smaller than the host genome (105 bp compared to 5x106 bp), replication of these factors to high copy number in the host places a considerable load on the host replication machinery. Naturally occuring origins of replication are therefore negatively regulated to keep copy number down (typically 5 to 10 copies per cell).
While high copy number is disadvantageous in a natural system, it is a desirable feature in a cloning vector - since the whole idea is to be able to easily isolate substantial quantities of particular DNA sequences. Therefore considerable work has gone into engineering the ColE1 ori such that the negative regualtory mechanisms that limit episome copy number are disabled. Modern plasmid vectors are therefore often called 'runaway replicons' and are present at 100 to 1000 copies per cell.
Selectable markers are essential for the identification of bacteria containing recombinant plasmids. Selection can be divided into two types:
Positive selection is used to identify bacteria that contain a plasmid. The most common markers used for positive selection are the antibiotic resistance genes carried by the original R factors.
While many antibiotics and resistance genes are available, the commonly used ones fall into two general classes:
Ampicillin is a beta-lactam based antibiotic that acts by inhibiting the synthesis of the bacterial peptidoglycan cell wall. Sensitive bacteria are not actively 'killed', but on cell division are unable to synthesize the cell wall and suffer from osmotic lysis.
The enzyme beta-lactamase is secreted into the periplasmic space where it breaks down the antibiotic, allowing cell wall synthesis to proceed.
The antibiotics tetracycline, kanamycin and chloramphenicol all act by inhibiting translation.
The covalent modification (phosphorylation, acetylation) of these antibiotics blocks their interaction with the translation apparatus.
The ligation of foreign DNA fragments into plasmid vectors is a relatively inefficient process - ligation can either produce recircularized plasmid with no insert, or plasmids containing a foreign DNA insert. These products are then transformed into an antibiotic sensitive bacterial host, an positive selection applied to identify bacteria that contain a plasmid. A second selection system is necessary to distinguish between plasmids that merely recircularized from those that carry a foreign DNA insert.
In order to identify those plasmids carrying a foreign DNA fragment, the site of insertion is chosen such that insertion disrupts a selectable marker - a phenomenon we call insertional inactivation.
Two types of selectable markers are used for negative selection
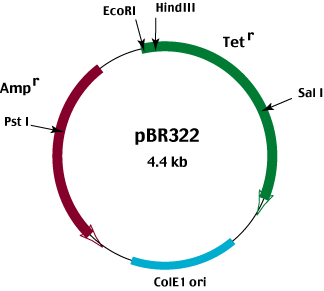
In order to use antibiotic resistance as a negative as well as a positive selection system, the plasmid vector must carry two different antibiotic resistance genes. An example of such a vector is pBR322.
pBR322 carries both an ampicllin and a tetracycline resistance gene.
The phenotype of bacteria containing the intact plasmid is Ampr Tetr
Insertion of foreign DNA into
the Pst I site located in the Ampr gene results in an Amps Tetr phenotype.
Conversely, insertion of foreign DNA into the EcoRI, Hind III or Sal I sites located in the Tetr gene results in an
Ampr Tets phenotype.
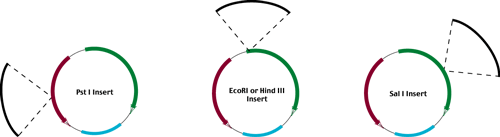
In order to reveal these phenotypes, transformants are first plated on positive selective media
(Tet or Amp respectively).
Positively selected colonies are then restreaked on positive selective media
(master plate from which we recover our desired vector + insert)
and on the negative selection media.
(Amp or Tet media respectively)
Ampr - Tets
Ampr - Tets
Amps - Tetr
Insertional Inactivation of Enzymatic Activity
While the insertional inactivation of antibiotic resistance works, it requires a lot of manipulation - picking the positively selected bacteria and replating on negative selection media etc. In addition to the tedium of picking colonies, vectors like pBR322 also suffer from a paucity of convenient restriction sites at which to insert the foreign DNA fragemnts.
These limitations promted the development of a set of host-vector systems in which it is possible to positively select for bacteria carrying a plasmid and simultaneously select for insertional inactivation of enzymatic activity. This system is based on our old friend, the
beta-galactosidase gene of the E coli lac operon.
go to
Plasmid Vectors II