Index
Lambda Biology
Libraries
Biology
Vectors
Vectors
In order to develop vectors that would carry larger foreign DNA inserts and to enable high density screens, we turn to bacteriophage lambda.
Lambda can adopt one of two alternate life styles on infection of its E coli host - lytic or lysogenic replication.
The initial stages of infection are the same for both life styles.
The infectious virion binds to the maltose receptor on the surface of the E coli. Binding induces a conformational change in the virion which expells the linear genome into the cell.
The ends of the linear molecule are called cos sites - cos for cohesive ends. In packaging the phage genome into the virion heads, the cos site - a 21 bp pallindromic sequence analogous to a large restriction site - in a staggered fashion generating single-stranded 5' overhangs.
On entering the host, these single stranded ends anneal to circularize the genome.
The lambda genome now becomes a template for transcription by the host RNA polymerase + sigma factor.
The immediate early phase of lambda genome transcription results in the transcription and translation of positive and negative transcription regulatory proteins.
The timing and relative abundance of these positive and negative factors produced during immediate early transcription results in the infection following the lytic or lysogenic pathway.
The delayed early phase is initiated by the action of these positive factors. The products of the delayed early transcription represent the replication functions of the phage.
Their expression initiates lambda genome replication.
Finally the late phase of infection produces the head and tail proteins of the phage virion.
These proteins self-0rganize to form the head capsid and tail fibre.
The head then packages the replicated copies of the lambda genome (in progress since the delayed early phase) and assembles with the tail fibre to form the infectious virion.
Phage lysis functions then lyse the host and release the progeny phage into the surrounding environment.
A single phage input replicates to produce about 1000 progeny in one cycle.
These thousand phage can then infect 1000 bacteria in the immediate environment.
One more lytic cycle and we have 1,000,000 progeny phage.
An efficient replication system eh?
The negative transcription regulator shuts off all lambda transcription except for its own expression.
With all lytic functions inaccessible, the lambda genome integrates into the host genome by a site-specific recombination event
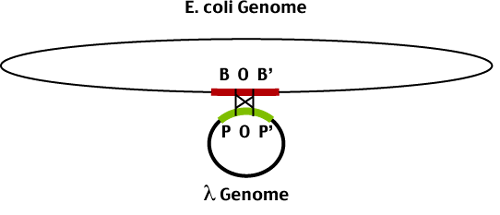
The site specific recombination occurs between two related sequences in the bacterial (BOB') and phage (POP') genomes. The central portion of these sequences (O) are identical and recombination takes place between them to produce the lysogen - an E coli genome containing an integrated phage genome. The phage genome is bounded by the recombinant joints (BOP' and POB').

While the bacterial host goes on with its busy life, the phage genome just goes along for the ride. The negative transcription factors continue to block phage genome transcription (except for their own expression). The phage genome replicates along with the host genome and is transmitted to all subsequent bacterial progeny of the original lysogenized cell.
It should be noted here that lysogeny in some obscure way does benefit the host. Once a bacterium has become a lysogen it is immune to subsequent infection. Any phage infecting a lysogenic cell is immediately shut down by the negative transcription regualtors being expressed from the integrated phage genome. But its the life of a ticking time bomb...
When life gets tough, the phage get going.
In particular, DNA damage (UV light etc) induces host DNA repair systems which inadvertantly results in the excision of the lambda lysogen. Host RecA protein destroys the lambda negative regulator and the lambda genome becomes a template for the expression of immediate early genes which lead to the excision of the prophage genome and entry into the lytic life cycle.
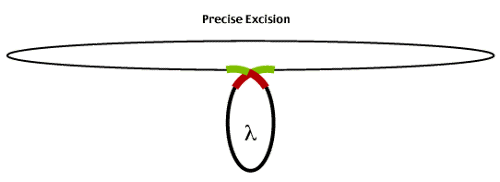
Infrequently, lamda excision produces phage that carry part of the E. coli genome flanking the integration site. These phage (called transducing phage as they 'transduce' E coli genomic sequences between hosts) result from imprecise excision events as illustrated below.

Imprecise excision starts with the alignment of BOP' and POB' but the resolution of the recombination event results in the exchange of sequences between the E. coli genome and the lambda genome. Notice that the phage contains BOP' and the E. coli genome POB' instead of the original BOB' and POP'. The phage sequences flanking the insertion site are replaced by E. coli genomic sequences and the corresponding portion of the lambda genome is left behind in the E. coli genome.
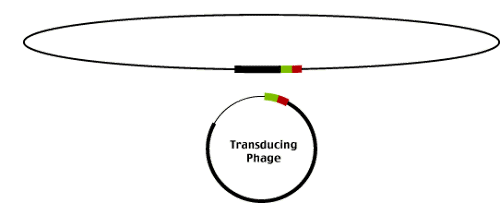
It was the identification of these transducing phage that first gave phage geneticists the idea that lambda might be useful as a cloning vector. Transducing phage already carry foreign DNA sequences (those derived from the E coli genome).
Lambda Vectors