DNA Structure
wrapped twice around a Histone octamer core.
Nucleosomes are linked together by approximately 50 bp of 'linker' DNA
to produce the beads-on-a-string appearance seen in electron micrographs as illustrated below.

Overall, packaging of DNA into nucleosomes results in a seven-fold compaction of the molecule's length
while increasing its diameter to approximately 10 nm (vs 2 nm for the naked helix).
Level III - The 30 nm Solenoid
The third level of hierarchical packaging involves the coiling of nucleosomes into a hollow tube (solenoid)
forming a fiber approximately 30 nm in diameter.
This level of condensation involves the interaction of a fifth Histone protein - Histone H1 - with the DNA
as it enters and exits the nucleosome via interaction with the two (H2A + H2B) dimers as shown below.
The compaction of nucleosomes into the 30 nm Solenoid involves not only Histone H1, but the 'unstructured' N-terminal tails of the core Histones which protrude out from the nucleosome core. These unstructured tails allow adjacent nucleosomes to interact to form the tightly packed solenoid structure observed in electron micrographs (we will discuss this interaction again when we talk about the regulation of gene expression).
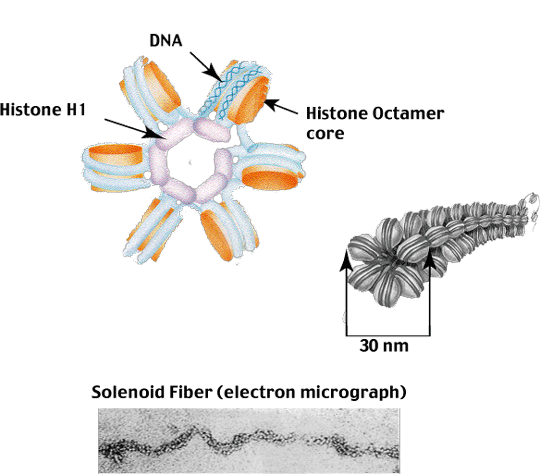
Packaging of nucleosomes into the 30 nm Solenoid results in approximately six-fold further compaction for a total of about 40-fold compaction of the naked DNA length.
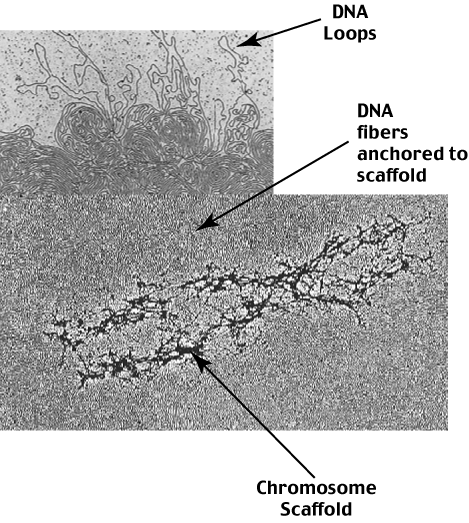
This level of compaction involves a new proteinaceous structure called the chromosome scaffold.
The scaffold itself is made up of several protein components including the lamins and topoisomerases.
The lamins are primarily structural while the topoisomerases are necessary for relieving supercoiling tension that builds up in the double-helix and ensures that the DNA molecule remains intact through the process of mitotic division.
The 30 nm solenoid is anchored to the chromosome scaffold at discrete sites called
Scaffold Attachment Regions (SARs)
and are organized into Radial Loop Domains of variable length.
Biology 335 Index